---
title: "4. Claret-Bruno model"
author:
- Daniel Sabanés Bové
- Francois Mercier
date: last-modified
editor_options:
chunk_output_type: inline
format:
html:
code-fold: show
html-math-method: mathjax
cache: true
---
This appendix shows how the Claret-Bruno model can be implemented in a Bayesian framework using the `brms` package in R.
## Setup and load data
{{< include _setup_and_load.qmd >}}
{{< include _load_data.qmd >}}
## Claret-Bruno model
In the Claret-Bruno model we have again the baseline SLD and the growth rate as in the Stein-Fojo model. Then in addition we have the inhibition response rate $\psi_{p}$ and the treatment resistance rate $\psi_{c}$. The model is then:
$$
y^{*}(t_{ij}) = \psi_{b_{0}i} \exp \left\{
\psi_{k_{g}i} t_{ij} -
\frac{\psi_{pi}}{\psi_{ci}} (1 - \exp(-\psi_{ci} t_{ij}))
\right\}
$$
for positive times $t_{ij}$. Again, if the time $t$ is negative, i.e. the treatment has not started yet, then it is reasonable to assume that the tumor cannot shrink yet. That is, we have then $\psi_{pi} = 0$. Therefore, the final model for the mean SLD is:
$$
y^{*}(t_{ij}) =
\begin{cases}
\psi_{b_{0}i} \exp(\psi_{k_{g}i} t_{ij}) & \text{if } t_{ij} < 0 \\
\psi_{b_{0}i} \exp \{
\psi_{k_{g}i} t_{ij} -
\frac{\psi_{pi}}{\psi_{ci}} (1 - \exp(-\psi_{ci} t_{ij}))
\} & \text{if } t_{ij} \geq 0
\end{cases}
$$
For the new model parameters we can again use log-normal prior distributions.
## Fit model
We can now fit the model using `brms`. The structure is determined by the model formula:
```{r}
#| label: specify_cb_model
formula <- bf(sld ~ ystar, nl = TRUE) +
# Define the mean for the likelihood
nlf(
ystar ~
int_step(year > 0) *
(b0 * exp(kg * year - (p / c) * (1 - exp(-c * year)))) +
int_step(year <= 0) *
(b0 * exp(kg * year))
) +
# Define the standard deviation (called sigma in brms) as a
# coefficient tau times the mean.
# sigma = tau * ystar is modelled on the log scale though, therefore:
nlf(sigma ~ log(tau) + log(ystar)) +
lf(tau ~ 1) +
# Define nonlinear parameter transformations
nlf(b0 ~ exp(lb0)) +
nlf(kg ~ exp(lkg)) +
nlf(p ~ exp(lp)) +
nlf(c ~ exp(lc)) +
# Define random effect structure
lf(lb0 ~ 1 + (1 | id)) +
lf(lkg ~ 1 + (1 | id)) +
lf(lp ~ 1 + (1 | id)) +
lf(lc ~ 1 + (1 | id))
# Define the priors
priors <- c(
prior(normal(log(65), 1), nlpar = "lb0"),
prior(normal(log(0.5), 0.1), nlpar = "lkg"),
prior(normal(0, 1), nlpar = "lp"),
prior(normal(log(0.5), 0.1), nlpar = "lc"),
prior(normal(2, 1), lb = 0, nlpar = "lb0", class = "sd"),
prior(normal(1, 1), lb = 0, nlpar = "lkg", class = "sd"),
prior(normal(0, 0.5), lb = 0, nlpar = "lp", class = "sd"),
prior(normal(0, 0.5), lb = 0, nlpar = "lc", class = "sd"),
prior(normal(0, 1), lb = 0, nlpar = "tau")
)
# Initial values to avoid problems at the beginning
n_patients <- nlevels(df$id)
inits <- list(
b_lb0 = array(3.61),
b_lkg = array(-0.69),
b_lp = array(0),
b_lc = array(-0.69),
sd_1 = array(0.5),
sd_2 = array(0.5),
sd_3 = array(0.1),
sd_4 = array(0.1),
b_tau = array(0.161),
z_1 = matrix(0, nrow = 1, ncol = n_patients),
z_2 = matrix(0, nrow = 1, ncol = n_patients),
z_3 = matrix(0, nrow = 1, ncol = n_patients),
z_4 = matrix(0, nrow = 1, ncol = n_patients)
)
# Fit the model
save_file <- here("session-tgi/cb3.RData")
if (file.exists(save_file)) {
load(save_file)
} else if (interactive()) {
fit <- brm(
formula = formula,
data = df,
prior = priors,
family = gaussian(),
init = rep(list(inits), CHAINS),
chains = CHAINS,
iter = WARMUP + ITER,
warmup = WARMUP,
seed = BAYES.SEED,
refresh = REFRESH,
adapt_delta = 0.9,
max_treedepth = 15
)
save(fit, file = save_file)
}
# Summarize the fit
save_fit_sum_file <- here("session-tgi/cb3_fit_sum.RData")
if (file.exists(save_fit_sum_file)) {
load(save_fit_sum_file)
} else {
fit_sum <- summary(fit)
save(fit_sum, file = save_fit_sum_file)
}
fit_sum
```
We did obtain here a warning about divergent transitions, see [stan documentation](http://mc-stan.org/misc/warnings.html#divergent-transitions-after-warmup) for details:
````
Warning message:
There were 4489 divergent transitions after warmup. Increasing adapt_delta above 0.9 may help. See http://mc-stan.org/misc/warnings.html#divergent-transitions-after-warmup
````
However, the effective sample size is high, i.e. the Rhat values are close to 1. This indicates that the chains have converged. We can proceed with the post-processing.
## Parameter estimates
```{r}
#| label: cb_post_processing
post_df_file <- here("session-tgi/cb3_post_df.RData")
if (file.exists(post_df_file)) {
load(post_df_file)
} else {
post_df <- as_draws_df(fit) |>
subset_draws(iteration = (1:1000) * 2)
save(post_df, file = post_df_file)
}
head(names(post_df), 10)
post_df <- post_df |>
mutate(
theta_b0 = exp(b_lb0_Intercept + sd_id__lb0_Intercept^2 / 2),
theta_kg = exp(b_lkg_Intercept + sd_id__lkg_Intercept^2 / 2),
theta_p = exp(b_lp_Intercept + sd_id__lp_Intercept^2 / 2),
theta_c = exp(b_lc_Intercept + sd_id__lc_Intercept^2 / 2),
omega_0 = sd_id__lb0_Intercept,
omega_g = sd_id__lkg_Intercept,
omega_p = sd_id__lp_Intercept,
omega_c = sd_id__lc_Intercept,
cv_0 = sqrt(exp(sd_id__lb0_Intercept^2) - 1),
cv_g = sqrt(exp(sd_id__lkg_Intercept^2) - 1),
cv_p = sqrt(exp(sd_id__lp_Intercept^2) - 1),
cv_c = sqrt(exp(sd_id__lc_Intercept^2) - 1),
sigma = b_tau_Intercept
)
```
Let's first look at the population level parameters:
```{r}
#| label: cb_pop_params
cb_pop_params <- c("theta_b0", "theta_kg", "theta_p", "theta_c", "sigma")
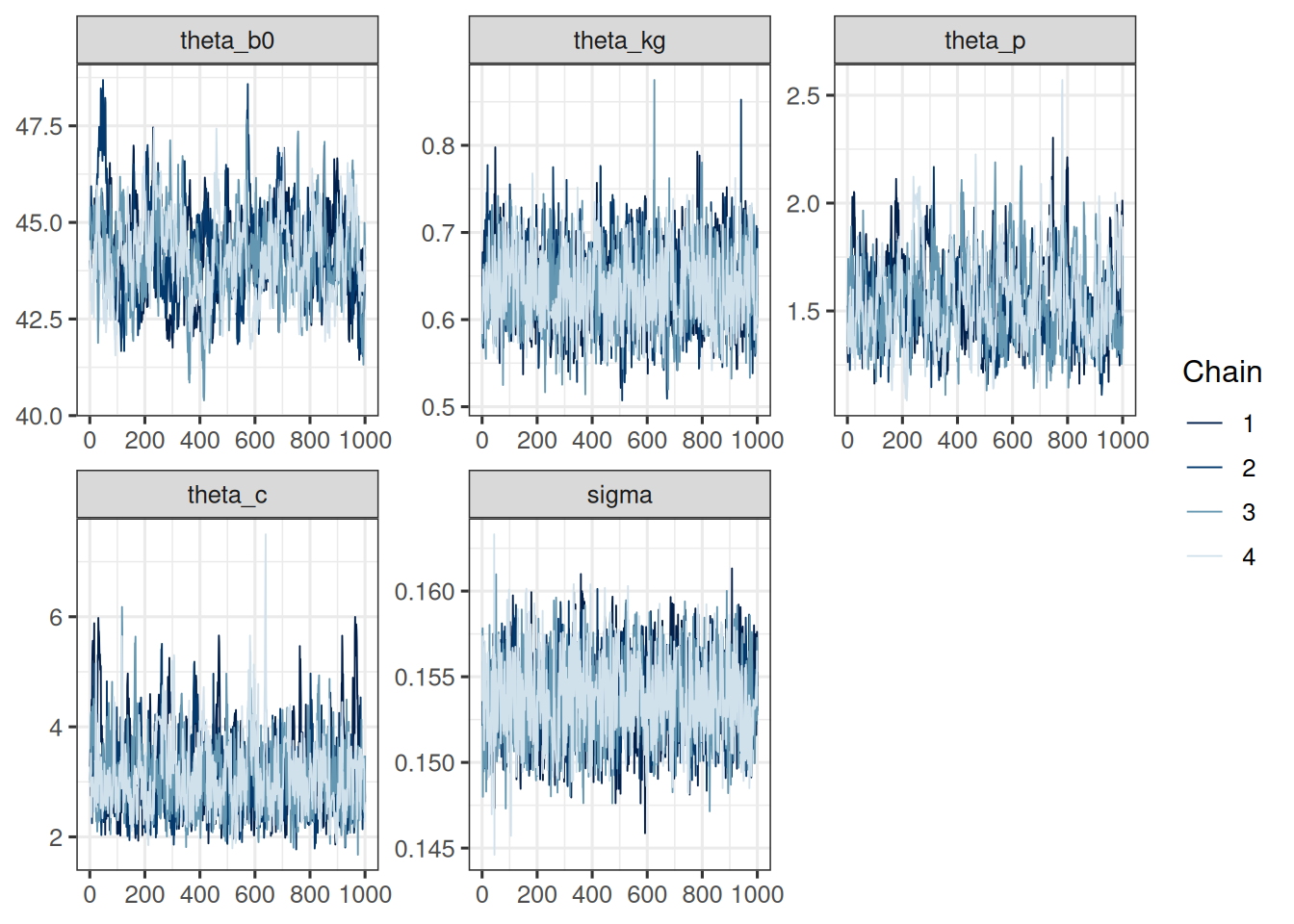
mcmc_trace(post_df, pars = cb_pop_params)
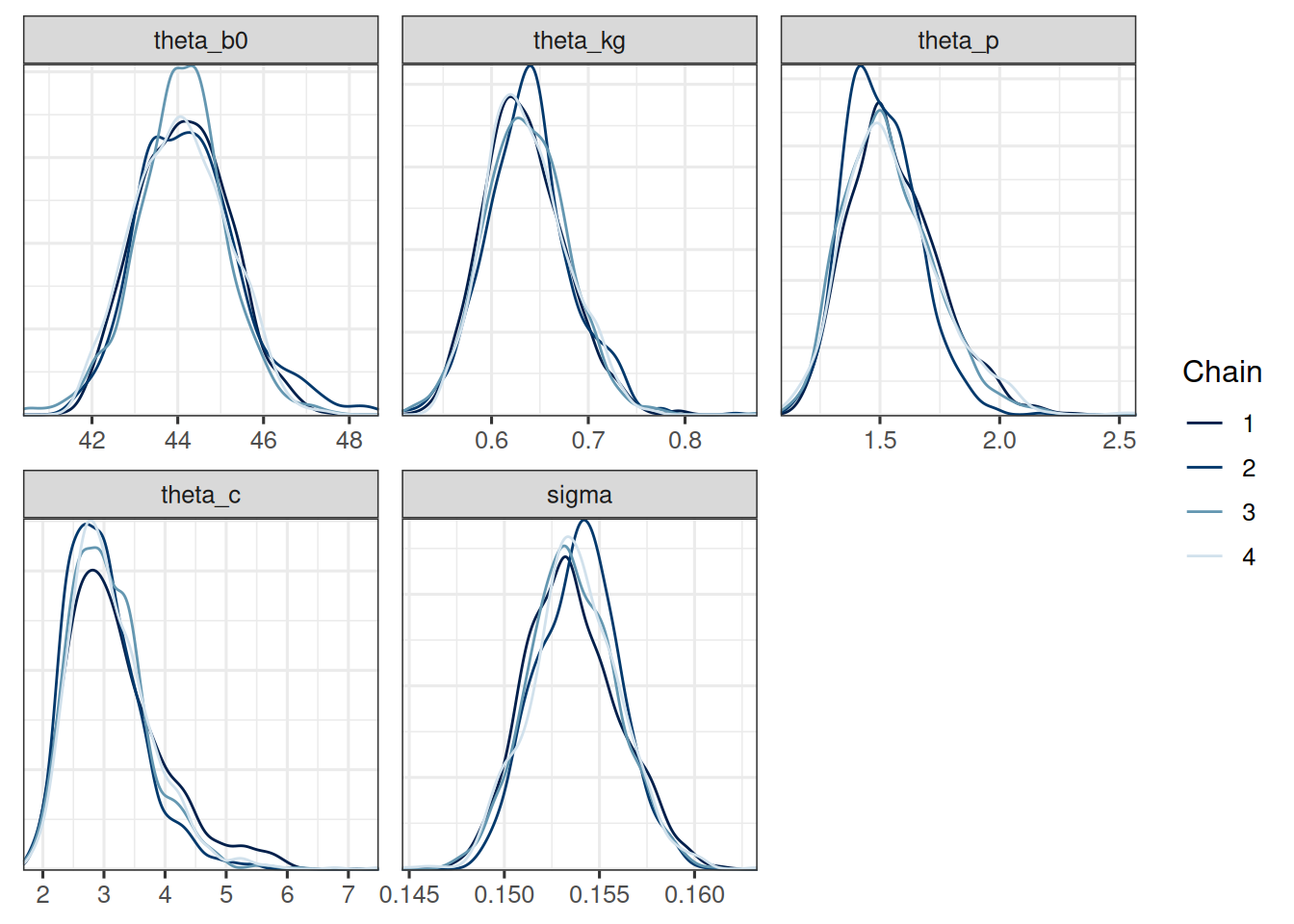
mcmc_dens_overlay(post_df, pars = cb_pop_params)
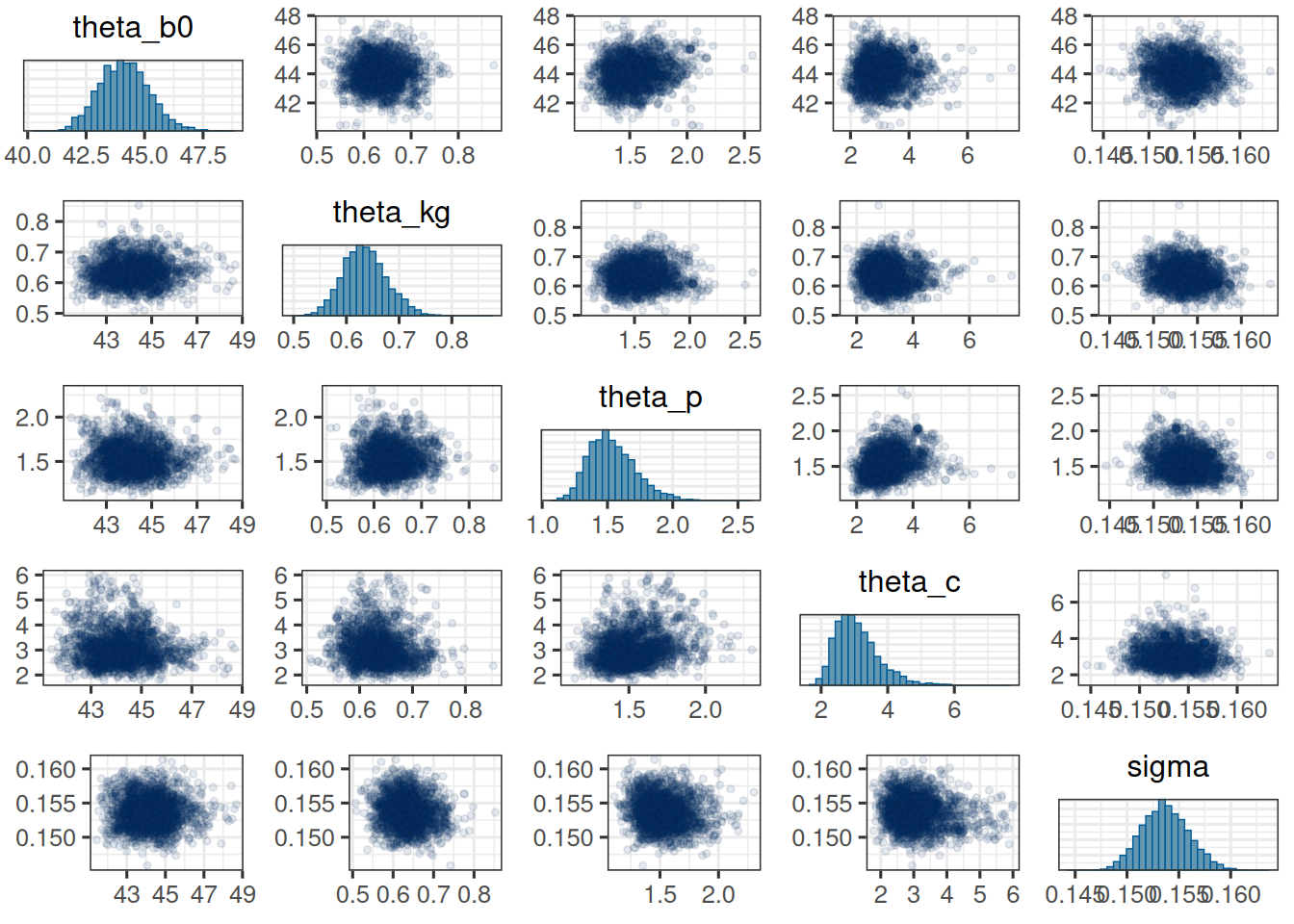
mcmc_pairs(
post_df,
pars = cb_pop_params,
off_diag_args = list(size = 1, alpha = 0.1)
)
```
The trace plots look good. The chains seem to have converged and the pairs plot shows no strong correlations between the parameters. Let's check the table:
```{r}
#| label: cb_post_summary
post_sum <- post_df |>
dplyr::select(theta_b0, theta_kg, theta_p, theta_c, omega_0, omega_g, omega_p, omega_c,
cv_0, cv_g, cv_p, cv_c,
sigma) |>
summarize_draws() |>
gt() |>
fmt_number(decimals = 3)
post_sum
```
We see similar estimated values as before for $\theta_{b_{0}}$, $\theta_{k_{g}}$ and $\sigma$.
## Observation vs model fit
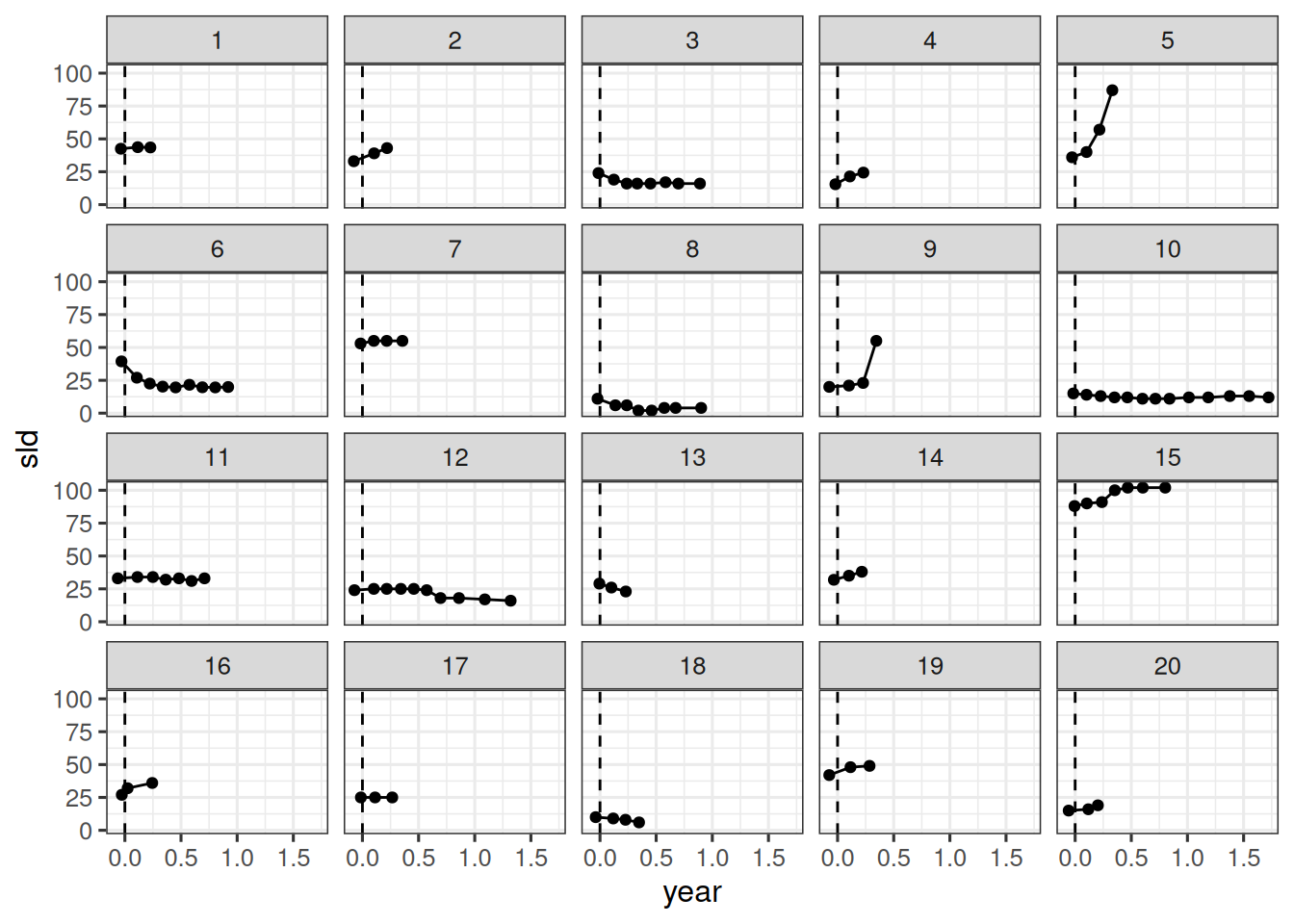
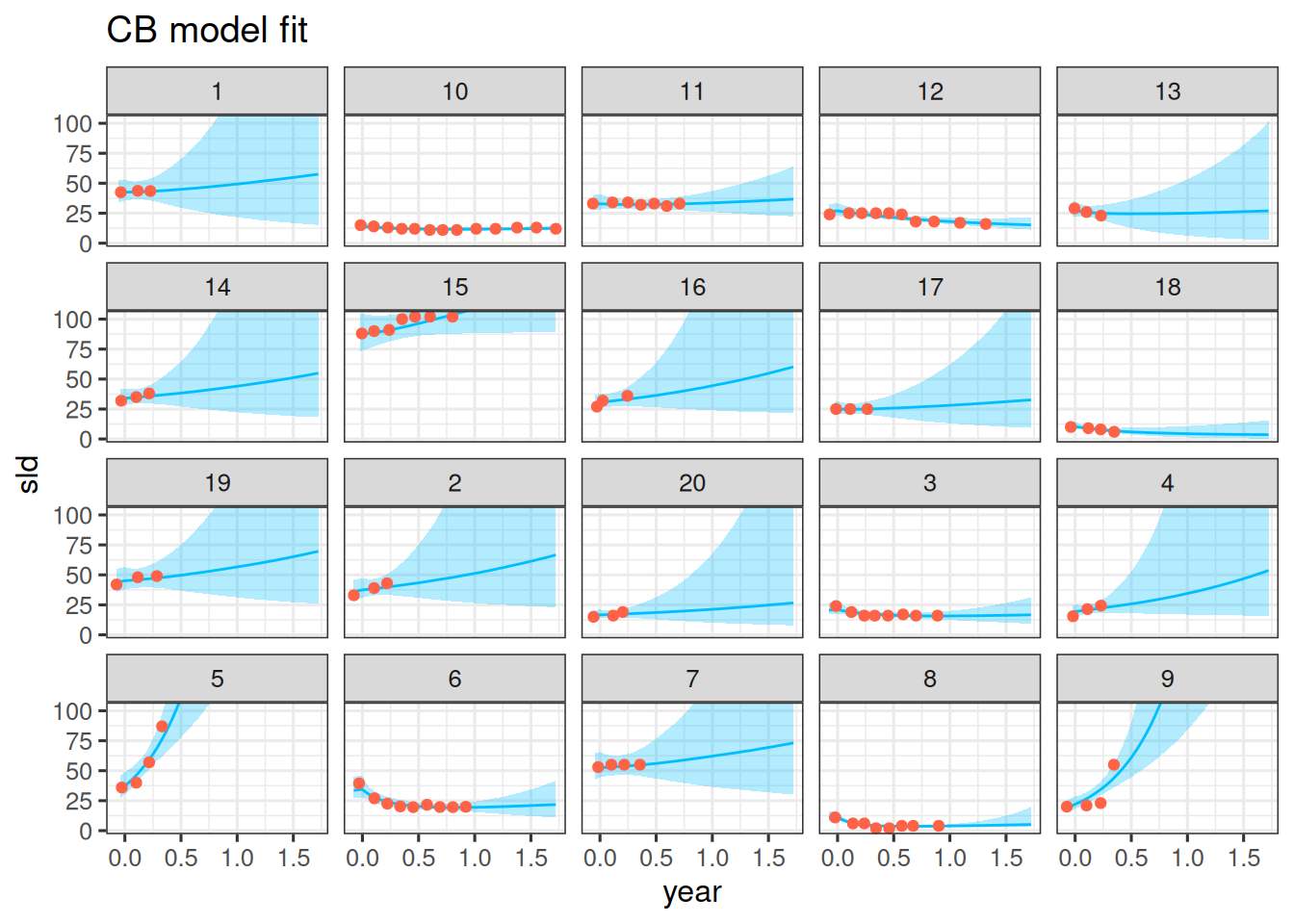
We can now compare the model fit to the observations. Let's do this for the first 20 patients again:
```{r}
#| label: cb_model_fit
pt_subset <- as.character(1:20)
df_subset <- df |>
filter(id %in% pt_subset)
df_sim_save_file <- here("session-tgi/cb3_sim_df.RData")
if (file.exists(df_sim_save_file)) {
load(df_sim_save_file)
} else {
df_sim <- df_subset |>
data_grid(
id = pt_subset,
year = seq_range(year, 101)
) |>
add_epred_draws(fit) |>
median_qi()
save(df_sim, file = df_sim_save_file)
}
df_sim |>
ggplot(aes(x = year, y = sld)) +
facet_wrap(~ id) +
geom_ribbon(
aes(y = .epred, ymin = .lower, ymax = .upper),
alpha = 0.3,
fill = "deepskyblue"
) +
geom_line(aes(y = .epred), color = "deepskyblue") +
geom_point(data = df_subset, color = "tomato") +
coord_cartesian(ylim = range(df_subset$sld)) +
scale_fill_brewer(palette = "Greys") +
labs(title = "CB model fit")
```
This also looks good. The model seems to capture the data well.
## With `jmpost`
This model can also be fit with the `jmpost` package. The corresponding function is `LongitudinalClaretBruno`. The statistical model is specified in the vignette [here](https://genentech.github.io/jmpost/main/articles/statistical-specification.html#claret-bruno-model).
Homework: Implement the generalized Claret-Bruno model with `jmpost` and compare the results with the `brms` implementation.