---
title: "2. OS model minimal workflow with `brms`"
author:
- Daniel Sabanés Bové
- Francois Mercier
date: last-modified
editor_options:
chunk_output_type: inline
format:
html:
code-fold: show
html-math-method: mathjax
cache: true
---
Let's try to fit the same model now with `brms`. We will use the same data as in the previous notebook.
## Setup and load data
```{r}
#| label: setup
#| echo: false
#| output: false
library(here)
options(knitr.duplicate.label = "allow")
knitr::purl(
here("session-os/_setup.qmd"),
output = here("session-os/_setup.R")
)
source(here("session-os/_setup.R"))
```
Here we directly start from the overall survival data with the log `kg` estimates, as we have obtained them in the previous notebook:
```{r}
os_data_with_log_kg <- readRDS(here("session-os/os_data_with_log_kg.rds"))
head(os_data_with_log_kg)
```
## Model fitting with `brms`
Let's first fit the model with `brms`. We will use the same (first) model as in the previous notebook, but we will use the `brms` package to fit it.
An important ingredient for the model formula is the censoring information, passed via the `cens()` syntax: This should point to a variable containing the value 0 for observed events, i.e. no censoring, and the value 1 for right censored times (see `?brmsformula` for more details). Therefore we first add such a variable to the data set:
```{r}
os_data_with_log_kg <- os_data_with_log_kg |>
mutate(
os_cens = ifelse(os_event, 0, 1)
)
```
We define our own design matrix with a column of ones:
```{r}
os_data_with_log_kg_design <- model.matrix(
~ os_time + os_cens + ecog + age + race + sex + log_kg_est,
data = os_data_with_log_kg
) |>
as.data.frame() |>
rename(ones = "(Intercept)")
head(os_data_with_log_kg_design)
```
Now we can define the model formula:
```{r}
formula <- bf(
os_time | cens(os_cens) ~
0 +
ones +
ecog1 +
age +
raceOTHER +
raceWHITE +
sexM +
log_kg_est
)
```
So here we suppress the automatic intercept provided by `brms` by using the `0 +` syntax, and instead we our "own" vector of ones. This is because we want to avoid the default centering of covariates which is performed by `brms` when using an automatic intercept. Otherwise it would be difficult to exactly match the prior distributions we used in the `jmpost` model further below.
In order to find out about the parametrization of the Weibull model with `brms` and Stan here, let's check the Stan code generated by `brms` for it:
```{r}
stancode(formula, data = os_data_with_log_kg_design, family = weibull())
```
We can see that the Stan code uses the `weibull_*` function family to define the log-likelihood contributions.
We can check the Stan reference doc [here](https://mc-stan.org/docs/functions-reference/positive_continuous_distributions.html#weibull-distribution) for details of the parametrization. We can see that this is the so-called "standard" parametrization (see [Wikipedia](https://en.wikipedia.org/wiki/Weibull_distribution#Standard_parameterization)) with shape parameter $\alpha$ and scale parameter $\sigma$. The mean of this distribution is $\sigma \Gamma(1 + 1/\alpha)$, where $\Gamma$ is the gamma function. We can see in the `brms` generated code that accordingly the `sigma` parameter is defined as `mu / tgamma(1 + 1 / shape)`, such that `mu` is really the mean of the distribution.
Now the problem is that this is a different parametrization than what we have used in `jmpost` (see the [specification](https://genentech.github.io/jmpost/main/articles/statistical-specification.html#weibull-distribution-proportional-hazard-parameterisation)), which is the proportional hazards parametrization (see [Wikipedia](https://en.wikipedia.org/wiki/Weibull_distribution#First_alternative)), where the covariate effects are on the log hazard scale instead of on the log mean scale. This has been identified by other `brms` users as a gap in the package, see e.g. [here](https://github.com/paul-buerkner/brms/issues/1677). So we can hope that this will be added in the future, but for now we need to implement a workaround.
Fortunately, we can define a custom distribution in `brms` to use the proportional hazards parametrization. This parametrization relates to the Stan Weibull density definition with the transformation of $\sigma := \gamma^{-1 / \alpha}$. The code here has been first written by Bjoern Holzhauer and was extended by Sebastian Weber to integrate more tightly with `brms` ([source](https://github.com/paul-buerkner/brms/issues/1677)). One thing to keep in mind here is that for technical reasons the first parameter of the custom distribution needs to be named `mu` and not `gamma`.
```{r}
family_weibull_ph <- function(link_gamma = "log", link_alpha = "log") {
brms::custom_family(
name = "weibull_ph",
# first param needs to be "mu" cannot be "gamma"; alpha is the shape:
dpars = c("mu", "alpha"),
links = c(link_gamma, link_alpha),
lb = c(0, 0),
# ub = c(NA, NA), # would be redundant
# no need for `vars` like for `cens`, brms can handle this.
type = "real",
loop = TRUE
)
}
sv_weibull_ph <- brms::stanvar(
name = "weibull_ph_stan_code",
scode = "
real weibull_ph_lpdf(real y, real mu, real alpha) {
// real sigma = pow(1 / mu, 1 / alpha);
real sigma = pow(mu, -1 * inv( alpha ));
return weibull_lpdf(y | alpha, sigma);
}
real weibull_ph_lccdf(real y, real mu, real alpha) {
real sigma = pow(mu, -1 * inv( alpha ));
return weibull_lccdf(y | alpha, sigma);
}
real weibull_ph_lcdf(real y, real mu, real alpha) {
real sigma = pow(mu, -1 * inv( alpha ));
return weibull_lcdf(y | alpha, sigma);
}
real weibull_ph_rng(real mu, real alpha) {
real sigma = pow(mu, -1 * inv( alpha ));
return weibull_rng(alpha, sigma);
}
",
block = "functions"
)
## R definitions of auxilary helper functions of brms, these are based
## on the respective weibull (internal) brms implementations:
log_lik_weibull_ph <- function(i, prep) {
shape <- get_dpar(prep, "alpha", i = i)
sigma <- get_dpar(prep, "mu", i = i)^(-1 / shape)
args <- list(shape = shape, scale = sigma)
out <- brms:::log_lik_censor(
dist = "weibull",
args = args,
i = i,
prep = prep
)
out <- brms:::log_lik_truncate(
out,
cdf = pweibull,
args = args,
i = i,
prep = prep
)
brms:::log_lik_weight(out, i = i, prep = prep)
}
posterior_predict_weibull_ph <- function(i, prep, ntrys = 5, ...) {
shape <- get_dpar(prep, "alpha", i = i)
sigma <- get_dpar(prep, "mu", i = i)^(-1 / shape)
brms:::rcontinuous(
n = prep$ndraws,
dist = "weibull",
shape = shape,
scale = sigma,
lb = prep$data$lb[i],
ub = prep$data$ub[i],
ntrys = ntrys
)
}
posterior_epred_weibull_ph <- function(prep) {
shape <- get_dpar(prep, "alpha")
sigma <- get_dpar(prep, "mu")^(-1 / shape)
sigma * gamma(1 + 1 / shape)
}
```
We can again check the Stan code that is generated for this custom distribution:
```{r}
stancode(
formula,
data = os_data_with_log_kg_design,
stanvars = sv_weibull_ph, # We pass the custom Stan functions' code here.
family = family_weibull_ph()
)
```
Indeed we can now use the custom distribution. We also see the default priors in the `transformed parameters` block on the shape parameter (`alpha`). We don't see an explicit prior on the regression coefficients (`b`), which means an improper flat prior is used by default.
The remaining challenge is that in `jmpost` we specified a Gamma prior for $\lambda$ which is now here the exponentiated intercept parameter. So in principle, we would need an `ExpGamma` prior on the intercept, meaning that if we exponentiate the intercept, it has a gamma distribution. However, this would again require a custom distribution. Let's try to go with an approximation: we can just draw samples from the `ExpGamma` distribution (by sampling from a gamma distribution and taking the log) and then approximate this with a skewed normal distribution (see [here](https://mc-stan.org/docs/functions-reference/unbounded_continuous_distributions.html#skew-normal-distribution) for the Stan documentation):
```{r}
set.seed(123)
intercept_samples <- log(rgamma(1000, 0.7, 1))
library(sn)
fit <- selm(intercept_samples ~ 1, family = "SN")
xi <- coef(fit, "DP")[1]
omega <- coef(fit, "DP")[2]
alpha <- coef(fit, "DP")[3]
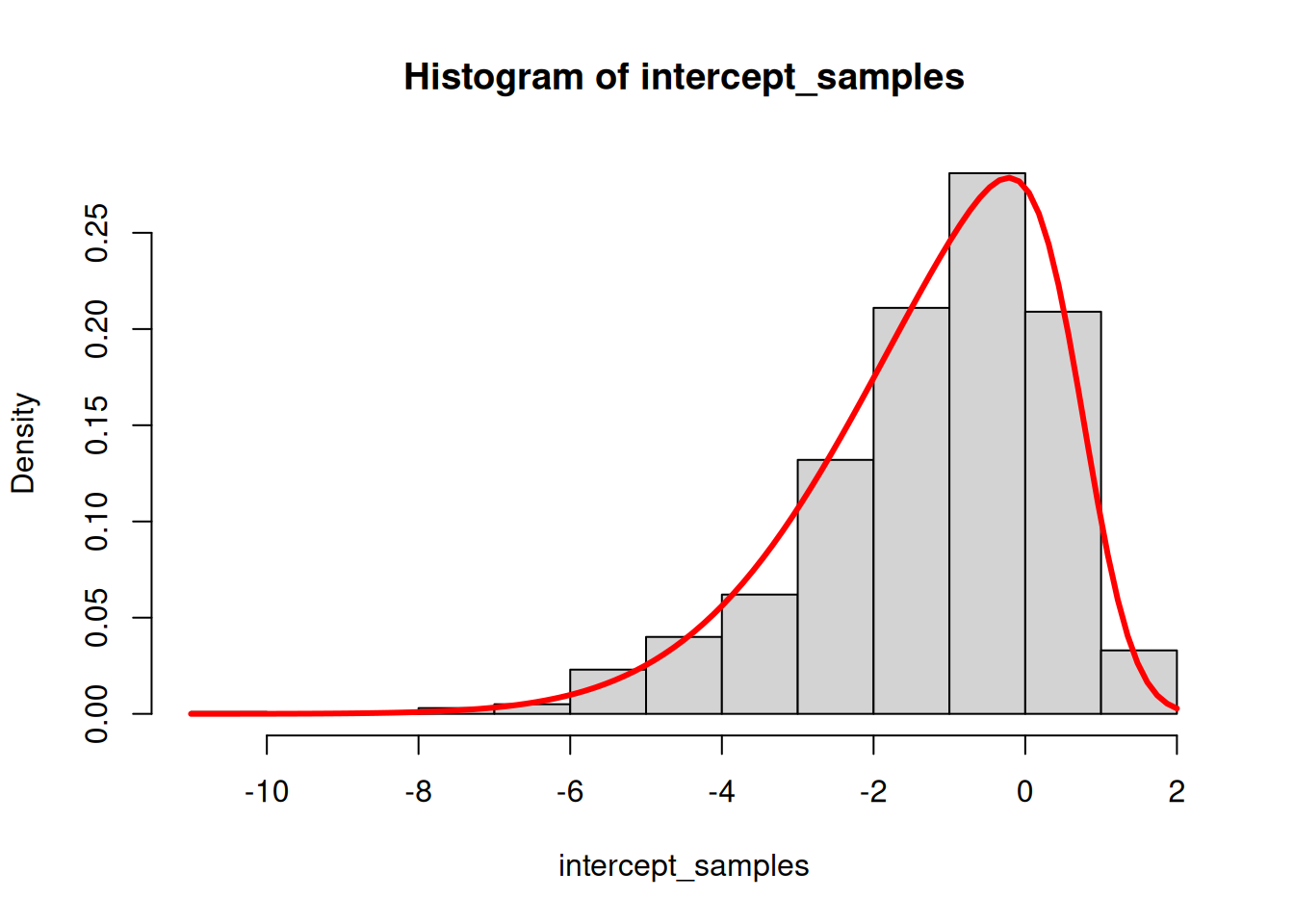
hist(intercept_samples, probability = TRUE)
curve(dsn(x, xi, omega, alpha), add = TRUE, col = "red", lwd = 3)
```
The skew normal density curve approximates the histogram of the log gamma samples well.
Now we can finally specify the priors:
```{r}
priors <- c(
set_prior(
glue::glue("skew_normal({xi}, {omega}, {alpha})"),
class = "b",
coef = "ones"
),
prior(normal(0, 20), class = "b"),
prior(gamma(0.7, 1), class = "alpha")
)
```
Let's do a final check of the Stan code:
```{r}
stancode(
formula,
data = os_data_with_log_kg_design,
prior = priors,
stanvars = sv_weibull_ph,
family = family_weibull_ph()
)
```
Now we can fit the model:
```{r}
save_file <- here("session-os/brms1.rds")
if (file.exists(save_file)) {
fit <- readRDS(save_file)
} else {
fit <- brm(
formula = formula,
data = os_data_with_log_kg_design,
prior = priors,
stanvars = sv_weibull_ph,
family = family_weibull_ph(),
chains = CHAINS,
iter = ITER + WARMUP,
warmup = WARMUP,
seed = BAYES.SEED,
refresh = REFRESH
)
saveRDS(fit, save_file)
}
summary(fit)
```
So the model converged fast and well.
## Comparison of results
Let's compare the results of the `brms` model with the `jmpost` model.
First we load again the `jmpost` results:
```{r}
draws_jmpost <- readRDS(here("session-os/os_draws.rds")) |>
rename_variables(
"gamma" = "sm_weibull_ph_gamma",
"lambda" = "sm_weibull_ph_lambda"
)
summary(draws_jmpost)
```
We prepare above `brms` results in the same format:
```{r}
draws_brms <- as_draws_array(fit) |>
mutate_variables(lambda = exp(b_ones)) |>
rename_variables(
"gamma" = "alpha",
"ecog1" = "b_ecog1",
"age" = "b_age",
"raceOTHER" = "b_raceOTHER",
"raceWHITE" = "b_raceWHITE",
"sexM" = "b_sexM",
"log_kg_est" = "b_log_kg_est"
) |>
subset_draws(
variable = c(
"ecog1",
"age",
"raceOTHER",
"raceWHITE",
"sexM",
"log_kg_est",
"gamma",
"lambda"
)
)
summary(draws_brms)
```
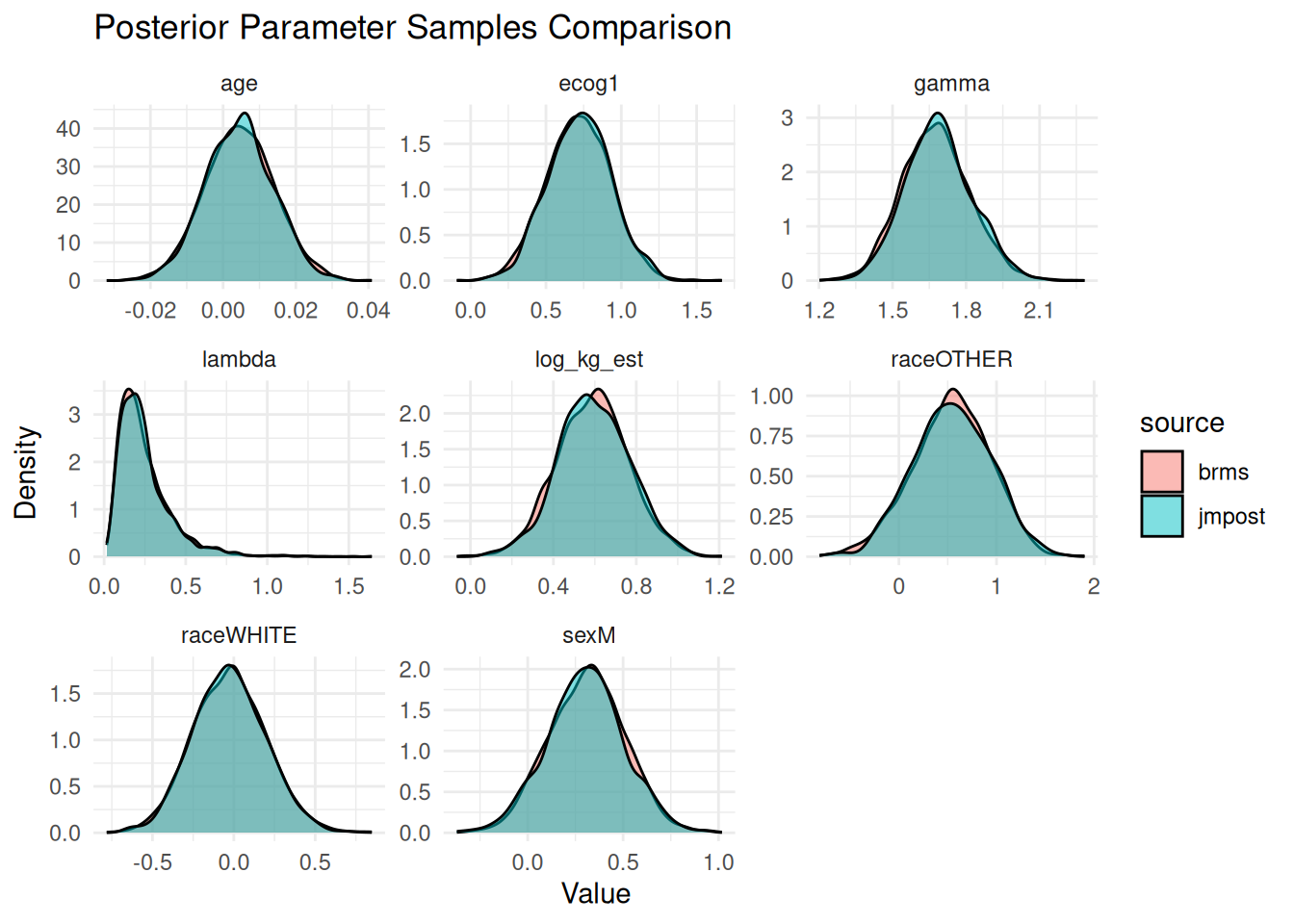
So the results agree well. We can also see this in density plots:
```{r}
# Combine the draws into one data frame
draws_combined <- bind_rows(
mutate(as_draws_df(draws_jmpost), source = "jmpost"),
mutate(as_draws_df(draws_brms), source = "brms")
) |>
select(-.chain, -.iteration, -.draw)
# Convert to long format for ggplot2
draws_long <- pivot_longer(
draws_combined,
cols = -source,
names_to = "parameter",
values_to = "value"
)
# Plot the densities
ggplot(draws_long, aes(x = value, fill = source)) +
geom_density(alpha = 0.5) +
facet_wrap(~parameter, scales = "free") +
theme_minimal() +
labs(
title = "Posterior Parameter Samples Comparison",
x = "Value",
y = "Density"
)
```
Generally these agree very well with each other. Overall we can expect a slight difference between the two results, because for the $\lambda$ parameter we only approximately used the same prior distribution in `brms` compared to `jmpost`.